WHAT DNA SAYS ABOUT THE FIRST HUMANS IN EAST ASIA
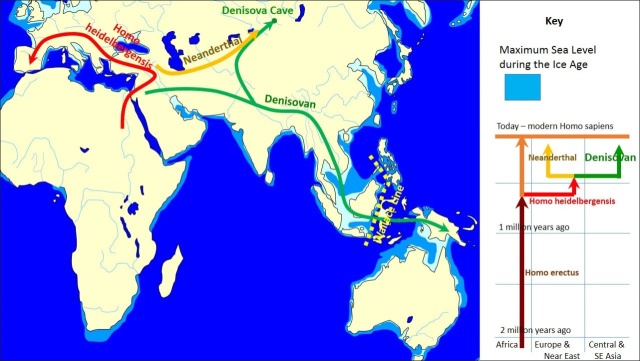
The first modern humans are believed to have arrived in eastern Asia in significant numbers about 60,000 to 40,000 years ago. Melinda A. Yang, a member of Dr. Qiaomei Fu’s Molecular Paleontology Lab at the Institute of Vertebrate Paleontology and Paleoanthropology, Chinese Academy of Sciences in Beijing, has been involved in extracting and sequencing ancient DNA using human remains from archaeological sites and using computational genomic tools to assess how their DNA related to that of previously sequenced ancient and present-day humans.
Yang wrote in The Conversation: One of our sequences came from ancient DNA extracted from the leg bones of the Tianyuan Man, a 40,000-year-old individual discovered near a famous paleoanthropological site in western Beijing. One of the earliest modern humans found in East Asia, his genetic sequence marks him as an early ancestor of today’s Asians and Native Americans. That he lived where China’s current capital stands indicates that the ancestors of today’s Asians began placing roots in East Asia as early as 40,000 years ago.[Source: Melinda A. Yang, Assistant Professor of Biology, University of Richmond, The Conversation, September 16, 2020]
“Farther south, two 8,000- to 4,000-year-old Southeast Asian hunter-gatherers from Laos and Malaysia associated with the Hòabìnhian culture have DNA that, like the Tianyuan Man, shows they’re early ancestors of Asians and Native Americans. These two came from a completely different lineage than the Tianyuan Man, which suggested that many genetically distinct populations occupied Asia in the past. But no humans today share the same genetic makeup as either Hòabìnhians or the Tianyuan Man, in both East and Southeast Asia. Why did ancestries that persisted for so long vanish from the gene pool of people alive now? Ancient farmers carry the key to that answer.
Based on plant remains found at archaeological sites, scientists know that people domesticated millet in northern China’s Yellow River region about 10,000 years ago. Around the same time, people in southern China’s Yangtze River region domesticated rice. Unlike in Europe, plant domestication began locally and was not introduced from elsewhere. The process took thousands of years, and societies in East Asia grew increasingly complex, with the rise of the first dynasties around 4,000 years ago. [Source: Melinda A. Yang, Assistant Professor of Biology, University of Richmond, The Conversation, September 16, 2020]
That’s also when rice cultivation appears to have spread from its origins to areas farther south, including lands that are today’s Southeast Asian countries. DNA helps tell the story. When rice farmers from southern China expanded southward, they introduced not only their farming technology but also their genetics to local populations of Southeast Asian hunter-gatherers. The overpowering influx of their DNA ended up swamping the local gene pool. Today, little trace of hunter-gatherer ancestry remains in the genes of people who live in Southeast Asia.
Farther north, a similar story played out. Ancient Siberian hunter-gatherers show little relationship with East Asians today, but later Siberian farmers are closely related to today’s East Asians. Farmers from northern China moved northward into Siberia bringing their DNA with them, leading to a sharp decrease in prevalence of the previous local hunter-gatherer ancestry.
RECOMMENDED BOOKS:
“Ancestral DNA, Human Origins, and Migrations” by Rene J. Herrera (2018) Amazon.com;
“Homo Sapiens Rediscovered: The Scientific Revolution Rewriting Our Origins” by Paul Pettitt Amazon.com;
“Who We Are and How We Got Here: Ancient DNA and the New Science of the Human Past” by David Reich (2019) Amazon.com;
“The Forgotten Exodus: The Into Africa Theory of Human Evolution” by Bruce Fenton (2017) Amazon.com;
“The Real Eve: Modern Man's Journey Out of Africa” by Stephen Oppenheimer (2004) Amazon.com;
“Asian Paleoanthropology: From Africa to China and Beyond” (Vertebrate Paleobiology and Paleoanthropology) by Christopher J. Norton and David R. Braun Amazon.com;
“Emergence and Diversity of Modern Human Behavior in Paleolithic Asia” by Yousuke Kaifu, Masami Izuho, et al. Amazon.com;
“Paleoanthropology and Paleolithic Archaeology in the People's Republic of China” by Wu Rukang, John W Olsen Amazon.com;
“In search of Homo erectus: a Prehistoric Investigation: The humans who lived two million years before the Neanderthals” by Christopher Seddon (2017) Amazon.com;
“Peking Man” Amazon.com;
“Java Man : How Two Geologists' Dramatic Discoveries Changed Our Understanding of the Evolutionary Path to Modern Humans” by Roger Lewin , Garniss H. Curtis, et al. Amazon.com;
“Little Species, Big Mystery: The Story of Homo Floresiensis” by Debbie Argue Amazon.com;
“Homo Floresiensis: Diving Deep into our Evolutionary Past” by Austin Mardon, Catherine Mardon, et al. Amazon.com;
“A New Human: The Startling Discovery and Strange Story of the "Hobbits" of Flores, Indonesia” by Mike Morwood, Penny van Oosterzee (2007) Amazon.com;
“Evolution: The Human Story” by Alice Roberts (2018) Amazon.com;
“Perspectives on Our Evolution from World Experts” edited by Sergio Almécija (2023) Amazon.com;
“Discovering Us: Fifty Great Discoveries in Human Origins” By Evan Hadingham (2021) Amazon.com;
“Our Human Story: Where We Come From and How We Evolved” By Louise Humphrey and Chris Stringer, (2018) Amazon.com;
“Lone Survivors: How We Came to Be the Only Humans on Earth” by Chris Stringer (2013) Amazon.com;
DNA Data from Asia
According to an article in Nature: The underrepresentation of non-Europeans in human genetic studies so far has limited the diversity of individuals in genomic datasets. Population-specific reference genome datasets as well as genome-wide association studies in diverse populations are needed to address this issue. A Korean reference genome as well as Japanese and Chinese reference genome datasets have been created. [Source: GenomeAsia 100K Project enables genetic discoveries across Asia, Nature, December 4, 2019]
For the GAsP project, we generated 1,267 high-coverage (average 36×) whole-genome sequences and analysed these together with 596 publicly available human genome sequences from previous sequencing studies. The 1,739 samples were enriched for individuals from population isolates to capture the broadest wealth of genetic diversity; the dataset includes 598 sequences from India, 156 from Malaysia, 152 from South Korea, 113 from Pakistan, 100 from Mongolia, 70 from China, 70 from Papua New Guinea, 68 from Indonesia, 52 from the Philippines, 35 from Japan and 32 from Russia .
In particular, India, Malaysia and Indonesia contain multiple ancestral populations as well as multiple admixed groups. On the basis of MSMC cross-coalescence rates, which reflect the increase in coalescence times of haplotypes sampled from different populations relative to haplotypes sampled from the same population, we estimate that the oldest population splits in southeast Asia and Oceania involve Melanesians and/or Negritos, who show a substructure from approximately 40 thousand years ago and evidence of separation around 20–30 thousand years ago. The population structure provides genetic information on classically defined population groups to aid future studies. For example, using multiple analytical approaches, we confirmed that the anthropologically classified ‘Negrito’ groups from India, Malaysia and the Philippines, are genetically more closely related to their geographical neighbours than they are to other Negrito groups, suggesting that dark skin colour is probably an environmental adaptation (for example, to high levels of solar radiation) and not an indicator of shared ancestry.
DNA Says Prehistoric Asians More Diverse Than Today’s Asians
Melinda A. Yang wrote Genetically speaking, today’s East Asians are not very different from each other. A lot of DNA is needed to start genetically distinguishing between people with different cultural histories. What surprised Dr. Fu and me was how different the DNA of various ancient populations were in China. We and others found shared DNA across the Yellow River region, a place important to the development of Chinese civilization. This shared DNA represents a northern East Asian ancestry, distinct from a southern East Asian ancestry we found in coastal southern China. [Source: Melinda A. Yang, Assistant Professor of Biology, University of Richmond, The Conversation, September 16, 2020]
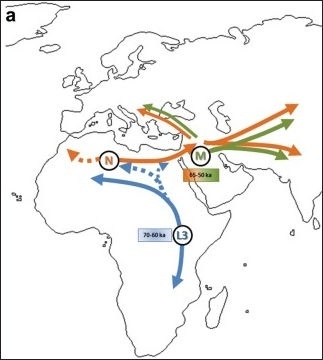
One possible model of L3's dispersal according to the Africa origin hypothesis: (a) Haplogroup N differentiates from L3 in the African continent, with a subsequent spread out of Africa.
“When we analyzed the DNA of people who lived in coastal southern China 9,000-8,500 years ago, we realized that already by then much of China shared a common heritage. Because their archaeology and morphology was different from that of the Yellow River farmers, we had thought these coastal people might come from a lineage not closely related to those first agricultural East Asians. Maybe this group’s ancestry would be similar to the Tianyuan Man or Hòabìnhians. But instead, every person we sampled was closely related to present-day East Asians. That means that by 9,000 years ago, DNA common to all present-day East Asians was widespread across China.
“Today’s northern and southern Chinese populations share more in common with ancient Yellow River populations than with ancient coastal southern Chinese. Thus, early Yellow River farmers migrated both north and south, contributing to the gene pool of humans across East and Southeast Asia. The coastal southern Chinese ancestry did not vanish, though. It persisted in small amounts and did increase in northern China’s Yellow River region over time. The influence of ancient southern East Asians is low on the mainland, but they had a huge impact elsewhere. On islands spanning from the Taiwan Strait to Polynesia live the Austronesians, best known for their seafaring. They possess the highest amount of southern East Asian ancestry today, highlighting their ancestry’s roots in coastal southern China. Other emerging genetic patterns show connections between Tibetans and ancient individuals from Mongolia and northern China, raising questions about the peopling of the Tibetan Plateau.
Andaman Islanders and Ancient Links to Africa and Asia
Nicholas Wade wrote in the New York Times: “Inhabitants of the Andaman Islands, a remote archipelago east of India, are direct descendants of the first modern humans to have inhabited Asia, geneticists conclude in a new study. But the islanders lack a distinctive genetic feature found among Australian aborigines, another early group to leave Africa, suggesting they were part of a separate exodus. [Source: Nicholas Wade, New York Times, December 10, 2002]
The Andaman Islanders are "arguably the most enigmatic people on our planet," a team of geneticists led by Dr. Erika Hagelberg of the University of Oslo write in the journal Current Biology. Their physical features — short stature, dark skin, peppercorn hair and large buttocks — are characteristic of African Pygmies. "They look like they belong in Africa, but here they are sitting in this island chain in the middle of the Indian Ocean," said Dr. Peter Underhill of Stanford University, a co-author of the new report. Adding to the puzzle is that their language, according to Joseph Greenberg, who, before his death in 2001, classified the world's languages, belongs to a family that includes those of Tasmania, Papua New Guinea and Melanesia.
Dr. Hagelberg has undertaken the first genetic analysis of the Andamanese with the help of two Indian colleagues who took blood samples — the islands belong to India — and by analyzing hair gathered almost a century ago by a British anthropologist, Alfred Radcliffe-Brown. The islands were isolated from the outside world until the British set up a penal colony there after the Indian mutiny of 1857.
Only four of the dozen tribes that once inhabited the island survive, with a total population of about 500 people. These include the Jarawa, who still live in the forest, and the Onge, who have been settled by the Indian government. Genetic analysis of mitochondrial DNA, a genetic element passed down only through women, shows that the Onge and Jarawa people belong to a lineage, known as M, that is common throughout Asia, the geneticists say. This establishes them as Asians, not Africans, among whom a different mitochondrial lineage, called L, is dominant.
The geneticists then looked at the Y chromosome, which is passed down only through men and often gives a more detailed picture of genetic history than the mitochondrial DNA. The Onge and Jarawa men turned out to carry a special change or mutation in the DNA of their Y chromosome that is thought to be indicative of the Paleolithic population of Asia, the hunters and gatherers who preceded the first human settlements.
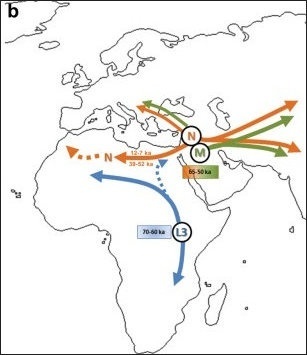
Another possible model of L3's dispersal according to the Africa origin hypothesis: b) Haplogroups M and N diverged from L3 outside Africa or during the expansion of L3-carrying AMH out of the continent; later migrations during Early Upper Paleolithic and the Neolithic diffusion led some lineages back to North Africa.
The mutation, known as Marker 174, occurs among ethnic groups at the periphery of Asia who avoided being swamped by the populations that spread after the agricultural revolution that occurred about 8,000 years ago. It is found in many Japanese, in the Tibetans of the Himalayas and among isolated people of Southeast Asia, like the Hmong. The discovery of Marker 174 among the Andamanese suggests that they too are part of this relict Paleolithic population, descended from the first modern humans to leave Africa.
Dr. Underhill, an expert on the genetic history of the Y chromosome, said the Paleolithic population of Asia might well have looked as African as the Onge and Jarawa do now, and that people with the appearance of present-day Asians might have emerged only later. It is also possible, he said, that their resemblance to African Pygmies is a human adaptation to living in forests that the two populations developed independently. A finding of particular interest is that the Andamanese do not carry another Y chromosome signature, known as Marker RPS4Y, that is common among Australian aborigines.
This suggests that there were at least two separate emigrations of modern humans from Africa, Dr. Underhill said. Both probably left northeast Africa by boat 40,000 or 50,000 years ago and pushed slowly along the coastlines of the Arabian Peninsula and India. No archaeological record of these epic journeys has been found, perhaps because the world's oceans were 120 meters lower during the last ice age and the evidence of early human passage is under water.
One group of emigrants that acquired the Marker 174 mutation reached Southeast Asia, including the Andaman islands, and then moved inland and north to Japan, in Dr. Underhill's reconstruction. A second group, carrying the Marker RPS4Y, took a different fork in Southeast Asia, continuing south toward Australia.
North-South Genetic Divide Among Native Populations in East Asia
Chinese researchers Feng Zhang, Bing Su, Ya-ping Zhang and Li Jin wrote in an article published by the Royal Society: In recent years researchers in China have made substantial efforts to collect samples and generate data especially for markers on Y chromosomes and mtDNA. The hallmark of these efforts is the discovery and confirmation of consistent distinction between northern and southern East Asian populations at genetic markers across the genome. With the confirmation of an African origin for East Asian populations and the observation of a dominating impact of the gene flow entering East Asia from the south in early human settlement, interpretation of the north–south division in this context poses the challenge to the field. Other areas of interest that have been studied include the gene flow between East Asia and its neighbouring regions (i.e. Central Asia, the Sub-continent, America and the Pacific Islands), the origin of Sino-Tibetan populations and expansion of the Chinese. [Source: “Genetic studies of human diversity in East Asia” by 1) Feng Zhang, Institute of Genetics, School of Life Sciences, Fudan University, 2) Bing Su, Laboratory of Cellular and Molecular Evolution, Kunming Institute of Zoology, 3) Ya-ping Zhang, Laboratory for Conservation and Utilization of Bio-resource, Yunnan University and 4) Li Jin, Institute of Genetics, School of Life Sciences, Fudan University. Author for correspondence (ljin007@gmail.com), 2007 The Royal Society, [Source: Genetic studies of human diversity in East Asia ncbi.nlm.nih.gov
***]Genetic markers are the tools in studying genetic variations. The most important genetic markers in human genetic diversity research (Du 2004) are: (I) blood groups that can be detected in red blood cells, including ABO, Rh and MNSs, (ii) human lymphocyte antigens and immunoglobulins, including Gm, Km and Am, (iii) isozyme markers, (iv) classic DNA polymorphisms using restriction fragment length polymorphism (RFLP), and (v) contemporary DNA markers, including short tandem repeat (STR or microsatellite) and single nucleotide polymorphisms (SNPs). However, it is the introduction of mtDNA and Y chromosome markers that has made a profound impact on our understanding of the genetic diversity of human populations . ***
Zhao et al. (1987) and Zhao & Lee (1989) studied the Gm and Km alleles (or allotypes) in 74 Chinese populations and found that there is an obvious genetic distinction between the southern and northern Chinese. By analysing a comprehensive dataset comprising 38 classical markers, Du & Xiao (Du et al. 1997) validated the genetic differentiation of southern and northern Chinese and showed that they are separated approximately by the Yangtze River. Chu et al. (1998) showed that such a north–south division can also be observed in Chinese populations using DNA markers (i.e. microsatellites). This genetic division is also consistent with multidisciplinary evidence in archaeology, physical anthropology, linguistics and surname distribution. Furthermore, the results from Chu et al. (1998) demonstrated that the north–south division is not limited to Chinese populations and is in fact a reflection of a north–south division of East Asian populations. In the last few years, genetic data on mtDNA and Y chromosomes have been accumulated at an unprecedented pace for East Asian populations. Again, a north–south division of East Asians was observed not only with Y chromosome markers . These observations provided convincing evidence of a north–south division in East Asian populations. ***
However, this well-established fact was not accepted without being challenged. Karafet et al. (2001) did not observe the north–south division in East Asians using a set of Y chromosome markers that are less polymorphic in East Asian populations and that over-represent the lineages brought in by recent admixture. In a different study, Ding et al. (2000) examined mtDNA, Y chromosome and autosomal variations and failed to observe a major north–south division. The southern populations in their study are primarily the Tibeto-Burman (TB) populations, which have a recent northern origin, and therefore would blur the north–south distinction . A more extensive study of mtDNA lineages provided a much higher resolution and consequently a strong north–south division emerged. ***
The north–south division raises the question of whether the southern and northern East Asians (NEAS) are descendants of the same ancestral population in East Asia or originated from different populations that arrived in East Asia via different routes. To date, three main hypotheses have been brought forward on the entry of modern humans into East Asia: (I) entry from Southeast Asia followed by northward migrations , (ii) entry from northern Asia followed by southward migrations , and (iii) southern and NEAS are derived from different ancestral populations, i.e. southern populations from Southeast Asia and northern populations from Central Asia. Therefore, to understand the mechanism of genesis and maintenance of the north–south division, much needs to be learnt about the origin and migration of the East Asians. ***
For the complete article from which the material here is derived see Genetic studies of human diversity in East Asia ncbi.nlm.nih.gov
DNA Says 'Denisovans' Roamed Widely In Asia
Denisovans are an extinct species or subspecies of archaic human that ranged across Asia during the Lower and Middle Paleolithic, and lived, based on current evidence, from 285,000 to 52,000 years ago. Named after the cave they were discovered in in Siberia, Denisovans have revealed few physical remains and thus most of what is known about them comes from DNA evidence. [Source: Wikipedia]
In 2010, AP reported, The Denisovan DNA code indicates they "roamed far from the cave that holds its only known remains. By comparing the DNA to that of modern populations, scientists found evidence that these "Denisovans" from more than 30,000 years ago ranged all across Asia. They apparently interbred with the ancestors of people now living in Melanesia, a group of islands northeast of Australia. There's no sign that Denisovans mingled with the ancestors of people now living in Eurasia, which made the connection between Siberia and distant Melanesia quite a shock.[Source: Malcolm Ritter, AP, December 22, 2010 ***]
"Scientists found evidence that in the genomes of people now living in Melanesia, about 5 percent of their DNA can be traced to Denisovans, a sign of ancient interbreeding that took researchers by surprise. "We thought it was a mistake when we first saw it," Reich said. "But it's real." And that suggests Denisovans once ranged widely across Asia, he said. Somehow, they or their ancestors had to encounter anatomically modern humans who started leaving Africa some 55,000 years ago and reached New Guinea by some 45,000 years ago. It seems implausible that this journey took a detour through southern Siberia without leaving a genetic legacy in other Eurasian populations, Reich said. It makes more sense that this encounter happened much farther south, indicating Denisovans ranged throughout Asia, over thousands of miles and different climate zones, he said. ***
Todd Disotell of New York University told AP he and colleagues were "blown away" by the unexpected Melanesia finding, with its implication for where Denisovans lived. "Clearly they had to have been very widespread in Asia," and DNA sampling of isolated Asian populations might turn up more of their genetic legacy, he said. Rick Potts, director of the human origins program at the Smithsonian Institution, said the new work greatly strengthens the case that Denisovans differed from Neanderthals and modern humans. Still, they may not be a new species, because they might represent a creature already known from fossils but which didn't leave any DNA to compare, such as a late-surviving Homo heidelbergensis, he said. ***
Potts also said the Melanesia finding could mean that the Melanesians and the Denisovans didn't intermix, but simply happened to retain ancestral DNA sequences that had been lost in other populations sampled in the study. But he stressed he doesn't know if that's a better explanation than the one offered by the authors. "I am excited about this paper (because) it just throws so much out there for contemplation that is testable," Potts said. "And that's good science." ***
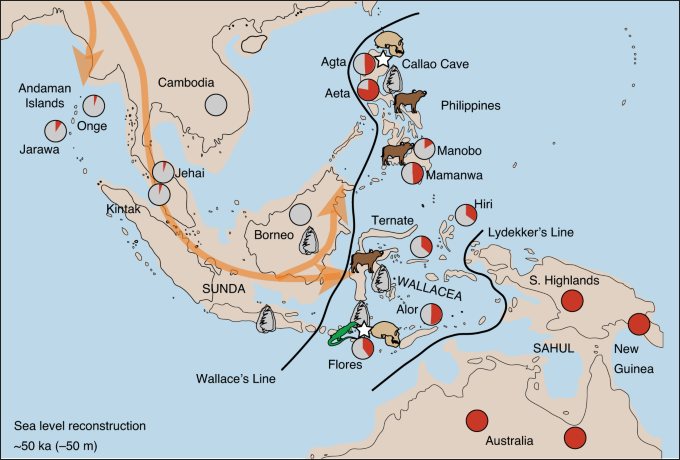
Where Denisovan DNA is highest (percentage in red in the circles)
Denisovan DNA in Asia
Our dense sampling of Asian populations enables the examination of Denisovan admixture in greater detail than has been previously possible, providing information about population splits or in-flows that occurred at or after the time of admixture (Supplementary Information 10). Our estimates of Denisovan ancestry were highest in Melanesians and the Aeta, intermediate in the Ati and groups from the Indonesian island of Flores, and low (but still significantly greater than 0) in most south, east and southeast Asian populations. We found high levels of Denisovan ancestry in Philippine Negrito groups but not in Malay or Andaman Negritos; these results are qualitatively similar to what was found in a previous study that was based on single-nucleotide polymorphism (SNP) arrays. The high levels of Denisovan ancestry in Melanesians and the Aeta are consistent with an admixture event into a population that is ancestral to both; however, two lines of evidence suggest that the ancestors of the Aeta experienced a second Denisovan admixture event. [Source: GenomeAsia 100K Project enables genetic discoveries across Asia, Nature, December 4, 2019]
First, multiple analyses found that the Aeta are genetically more similar to populations without appreciable Denisovan ancestry (for example, Igorot, Malay and Malay Negrito groups) than they are to Melanesians. This can be explained by more recent gene flow from other populations without Denisovan ancestry. However, such gene flow would reduce the levels of Denisovan admixture below that found in Melanesians. More directly, we find that putative Denisovan haplotypes that are unique to the Aeta (n = 962) are significantly longer than putative Denisovan haplotypes shared between Aeta and Papuans (n = 596, mean = 16.1 kb compared with mean = 14.1 kb, Mann–Whitney U-test, P ≪ 10−10), or putative Denisovan haplotypes unique to Papuans (n = 727, mean = 16.1 kb compared with mean = 14.9 kb, Mann–Whitney U-test, P ≪ 10−1,000), supporting a scenario in which a second admixture event between the Aeta and Denisovans happened after the separation of the Aeta and Melanesians. Two distinct Denisovan admixture events are most consistent with Homo sapiens and Denisovans interacting within southeast Asia, making it likely that admixture occurred within Sundaland or even farther east.
“A recent study found a slightly increased amount of Denisovan ancestry in south Asians compared with a priori expectations. We examined whether this was correlated with either language or social and/or caste status. South Asian samples were grouped into individuals who speak Indo-European languages and individuals who speak non-Indo-European languages (excluding individuals who speak Tibeto-Burman languages), as well as four social or cultural groups: tribal (Adivasi) groups, lower-caste groups, high-caste groups and Pakistani groups (Indo-European language speaking only). We found that the average levels of Denisovan ancestry were significantly different between the four social or cultural groups. Our results are consistent with the scenario that Indo-European-speaking migrants who entered the subcontinent from the northwest admixed with an indigenous South Asian (ancestral south Indian) group who had higher levels of Denisovan ancestry.

Migration of haplogroups ADN-Y
DNA Haplogroups and the Migration from Africa to Asia
Aileen Kawagoe wrote: All people living today can trace their maternal ancestry back to one of 26 core mtDNA (mitochondrial DNA) Haplogroups. Haplogroups are the main “trunks” of the mtDNA phylogenetic tree and represent extremely ancient family groups which arose tens of thousands of years ago. Over time, the descendents of each Haplogroup formed further subgroups, called “Subclades”. By discovering which Haplogroup “family” you belong to, you can then trace even further which sub-branch of your Haplogroup you belong to through “Subclade” analysis.[Source: Aileen Kawagoe, Heritage of Japan website, heritageofjapan.wordpress.com]
“Modern humans left Africa, moving along the tropical coastlines (and the earliest modern human fossil found outside of Africa was) about 100,000 years ago. Hugging the coastline, the early modern human was probably able to expand and occupy or colonize coastal areas by exploiting marine food resources by that time. The modern humans were thought to have been possibly forced out of Africa due to the East African mega-droughts (about 135,750 years ago) during the early late-Pleistocene. The period between 80,000-10,000 years ago during the last glacial would have had a huge impact on modern human migration, and the sea level had fallen 50-200 meters below present, which resulted in larger dry lands and possibility for human migration between currently separated lands by ocean, e.g. between Japan and the mainland.
“The new arrivals bore lineages (the YAP/M145/M203/SRY4064) that are traceable to an African origin, as the M-174 ancestral allele among YAP+ve lineages is found exclusively in Africa. (M-168 lineages evolved into three distinct sub-clusters: YAP at DYS287 as well as the M-145 and M-203, and two other lineages, defined by the distinct mutations RPS4Y/M216 and M89/M213. These three sub-clusters represent deep structuring of Y-chromosome diversity outside Africa – out of these lineages.) The subcluster found in East Asia is defined and identified by the M-174 mutation.
“Around 60,000 years ago, the first modern humans very likely arrived in Southern East Asia after having proceeded via the Indian sub-continental coastline. These earliest of migrating peoples were the bearers of the Haplogroup types C and D. These explorers originally of African origin settled initially in mainland southern East Asia, then migrated northward about 25,000-30,000 years ago, and spread throughout East Asia.
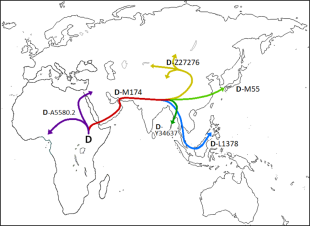
Haplogroup D in Asia
Aileen Kawagoe wrote: “Haplogroup D is believed to represent the Great Coastal Migration along southern Asia, from Arabia to Southeast Asia and thence a northward migration to populate East Asia. It is found today at high frequency among populations in Tibet, the Japanese archipelago, and the Andaman Islands, though curiously not in India. It is found at especially high frequencies among Andaman Islanders and 8-65 percent in northeast Indian tribes.[Source: Aileen Kawagoe, Heritage of Japan website, heritageofjapan.wordpress.com]
“However, the fragmented distribution of one East Asian specific Y chromosome lineage – D-M174, which is found at high frequencies only in Tibet, Japan and the Andaman Islands, is inconsistent with this scenario. The Ainu of Japan and the Jarawa and Onge of the Andaman Islands are notable for possessing almost exclusively Haplogroup D chromosomes (although Haplogroup C3 chromosomes also occur among the Ainu at a frequency of approximately 15 percent). Haplogroup D chromosomes are also found at low to moderate frequencies among populations of Central Asia and northern East Asia as well as the Han and Miao-Yao peoples of China and among several minority populations of Sichuan and Yunnan that speak Tibeto-Burman languages and reside in close proximity to the Tibetans. Also, the Haplogroup D Y-chromosomes that are found among populations of the Japanese Archipelago are particularly distinctive, bearing a complex of at least five individual mutations along an internal branch of the Haplogroup D phylogeny, thus distinguishing them clearly from the Haplogroup D chromosomes that are found among the Tibetans and Andaman Islanders.
“Haplogroup D is believed to have originated in Asia some 60,000 years before present…but where in Asia? While haplogroup D along with haplogroup E contains the distinctive YAP polymorphism (which indicates their common ancestry), no haplogroup D chromosomes have been found anywhere outside of Asia. Haplogroup D is also remarkable for its rather extreme geographic differentiation, with a distinct subset of Haplogroup D chromosomes being found exclusively in each of the populations that contains a large percentage of individuals whose Y-chromosomes belong to Haplogroup D: Haplogroup D1 among the Tibetans (as well as among the mainland East Asian populations that display very low frequencies of Haplogroup D Y-chromosomes), Haplogroup D2 among the various populations of the Japanese Archipelago, Haplogroup D3 among the inhabitants of Tibet, Tajikistan and other parts of mountainous southern Central Asia, and paragroup D* (probably another monophyletic branch of Haplogroup D) among the Andaman Islanders. Another type (or types) of paragroup D* is found at a very low frequency among the Turkic and Mongolic populations of Central Asia, amounting to no more than 1 percent in total. This apparently ancient diversification of Haplogroup D suggests that it may perhaps be better characterized as a “super-haplogroup” or “macro-haplogroup.” In one study, the frequency of haplogroup D* found among Thais was 10 percent.
“Unlike haplogroup C, Haplogroup D carriers most likely did not travel from Asia to the New World — it is found in no modern Native American (North, Central or South) populations (it is however not impossible that Haplogroup D may have traveled to the New World, like Haplogroup C, but that those lineages had died out).
Asian Groups with a High For Percentage of Haplogroup D
Aileen Kawagoe wrote: Besides Tibetans and Japanese, D-M174 is also prevalent in several southern ethnic populations in East Asia, including the Tibeto-Burman speaking populations from Yunnan province of southwestern China (14.0–72.3 percent), one Hmong-Mien population from Guangxi of southern China (30 percent) and one Daic population from Thailand (10 percent), which could be explained by fairly recent population admixture. However, a recent study reported a high frequency of D-M174 in Andamanese (56.25 percent), people who live in the remote islands in the Indian Ocean and considered one of the earliest modern human settlers of African origin in Southeast Asia. Another study suggested that the D-M174 lineage likely reached East Asia about 50,000 years ago. This implies that the YAP lineage in East Asia could be indeed very ancient.
“Sub-haplogroup D-M174 is now widely held to have a Tibetan or south China origin. D1 (M15) is found at high frequencies among Qiang people, 30 percent with a moderate distribution throughout East Asia; D2 (M55) is found at high frequencies among the Ainu, Japanese, and Ryukyuans. Frequencies: Ainu 87 percent, Okinawa 56 percent, Honshu 37 percent and Kyushu28 percent.; D3a (P47) Found at high frequencies among Tibetans among the Pumi people at 70 percent, with a moderate distribution among some other populations of southern Central Asia. The D2 subclade (SNP P37.1 and M55) is very rare outside of Japan and not found in Tibet, the only other region that has substantial amounts of the D haplogroup. Scientists’ calculations show a D2 population founding at 20-37,ooo years ago and an expansion that took place around 12,500 years ago. Subclade descendants D2a (SNP M116.1), D2a1 (SNP M125) and D2a1b (SNP 022457) are also Japan-specific. The long diversified branch of Haplogroup D found in Japan fits a model of genetic isolation for a long period of time.
Spread of Haplogroups from Southeast Asia to North Asia and Japan
Aileen Kawagoe wrote: “Austro-Asiatic homeland of populations bearers of C and D clades originate possibly around a Daic-speaking source in the mainland around South China and Vietnam (adjacent to the Gulf of Tonkin). Because most Austro-Asiatic populations have significant levels of Haplogroup C (and other old lineages M1 and K) there is good reason to believe that this language group also has an ancient origin, despite their recent contribution to populations in Oceania. A case of limited gene flow supports the spread of Austronesian language through the South Pacific more by cultural diffusion than through demic diffusion (i.e. culture and language is likely to be spread more rapidly than genes). [Source: Aileen Kawagoe, Heritage of Japan website, heritageofjapan.wordpress.com]
“Recent genetic studies have shown that 80 percent of Taiwan’s population also have Austro-Asiatic ancestry. The Taiwanese aborigines also share genetic heritage and the Austronesian language with SEAS populations and parts of Melanesia. However, it appears that the spread of this language group did not hop from Taiwan to Melanesia, but rather diffused separately to both regions from a Daic-speaking source in the mainland around South China and Vietnam (adjacent to the Gulf of Tonkin). The human males moving further east into remote Polynesia, therefore, appear to have combined origins from Austro-Asiatic/Austronesian (SEAS) and non-Austronesian (Melanesia) populations.
“At around 20,000 – 30,000 years ago, the Southern Eastern Asian/Austronesian populations carrying Haplogroup clades C and D migrated northward, spreading throughout East Asia. A recent archaeological finding supported that modern humans explored the Tibetan plateau about 30,000–40,000 years ago, which is much earlier than the conservative estimates of 20-30,000 years ago.
“Meanwhile, a haplogroup subclade D-carrying population enters Japan. As mentioned earlier, Haplogroup D is thought to be a remnant of more ancient population foundings, to constitute tribes representing relic traces of Haplogroup D connections between Tibet or south China and Japan. The post-glacial sea level rise eventually led to the separation between Japan and the main continent, which explains the relic distribution of D-M174 in current Japanese populations. The archaeological data suggested that the initial colonization of modern humans in Japan occurred about 30,000 years ago, consistent with the age estimation of D2-M57 (37,678 ± 2,216 years ago). Around 20-35,000 years ago, a second migration was made by people — involving carriers of Haplogroup clades O, N and P. The above population movements taken into consideration are the basis of current theories that the current Tibetan and Japanese populations are probably the admixture of two ancient populations represented by D-M174 and O3-M122 respectively.
Image Sources: Wikimedia Commons except 2nd Denisovan map from Nature
Text Sources: National Geographic, New York Times, Washington Post, Los Angeles Times, Smithsonian magazine, Nature, Scientific American. Live Science, Discover magazine, Discovery News, Natural History magazine, Archaeology magazine, The New Yorker, Time, BBC, The Guardian, Reuters, AP, AFP and various books and other publications.
Last updated April 2024